During the past months, the cBioPortal community has worked on facilitating the setup of local instances using Docker. You can read how to create your own in our article. Once this is ready, new public or private studies can be loaded into cBioPortal. The cBioPortal Datahub has ready-to-load files for more than 350 public studies.
In this aricle, we will show all the steps to load a new study in cBioPortal.
Prerequisites
As mentioned, uploading new studies depends on having a healthy local cBioPortal instance. Please make sure that the relevant containers are running: cBioPortal, MySQL, MongoDB and session service.
Steps
(1) Downloading the data: We clearly need data files that can be uploaded into the portal. For this example, we will use the TCGA Bladder Cancer study. You can download it directly from the repository using this link or using git lfs. If using git lfs, please first install lfs. Then, clone the repository and pull your chosen study:
git lfs install --skip-repo --skip-smudge
git clone https://github.com/cBioPortal/datahub.git
cd dathub
git lfs install --local --skip-smudge
Git lfs pull -I public/blca_tcga
(2) Running commands inside a container is not too different from running a command in a command line. Here we run the main command metaImport.py from the container running the cbioportal image.
docker-compose run \
-v “<path-to-datahub-folder>:/study” \
-v “<path-to-validation-reports>:/report”\
cbioportal \
metaImport.py \
-u http://cbioportal:8080 \
-s /study/blca_tcga \
--html=/report/report.html
We need to mount the volumes for the study data, and a directory where the validation reports will be stored. Besides that, the metaImport.py script requires the parameters -u (your local address), -s (the path to the study) and --html (the name of the validation report that will be created).
The metaImport.py script runs two main processes. First, it runs the validation for the study files. The validation checks that the data follow the cBioPortal format and can be imported. An html validation report is saved in the path set by the - -html flag. Once it is completed, the importing will begin. This incorporates the data into the MySQL database to be read by the portal. If the study is loaded successfully, the end of the script will output a message: “Updating study status to : 'AVAILABLE' for study: blca_tcga”.
Last steps: Before the study is accessible from the portal, we need to restart Tomcat to retrieve any updates from the database by simply restarting the cbioportal container:
docker-compose restart cbioportal
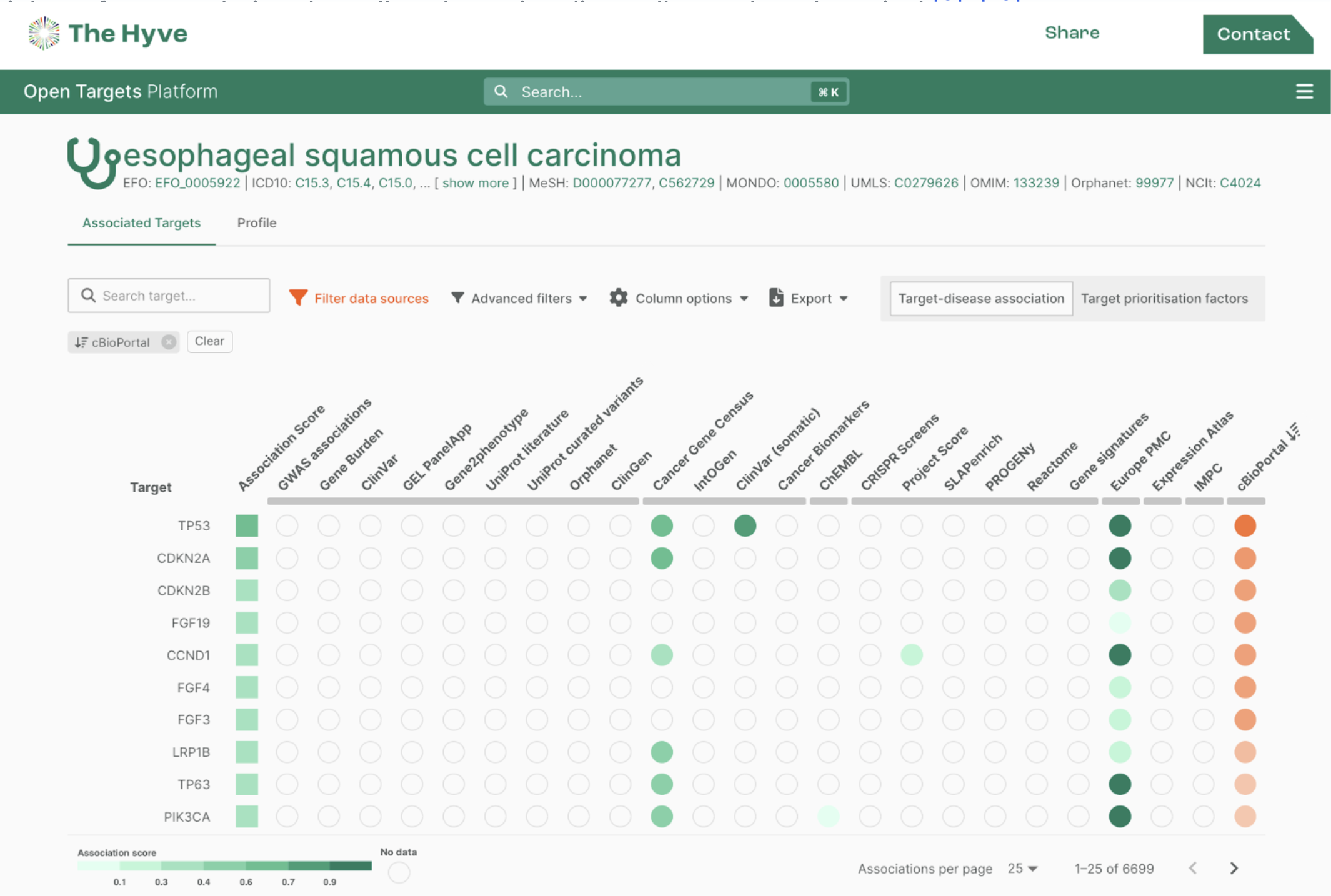
Finally, you can visit your study in your local cBioPortal instance!
Additional functionality
Having a local instance allows you to load complementary files, such as gene panels and gene sets, or to remove studies. This Docker strategy can be used in a similar way to run those commands.
More information about the importer script can be found in the cBioPortal documentation. For commercial support with data loading or transformation, you can contact us.