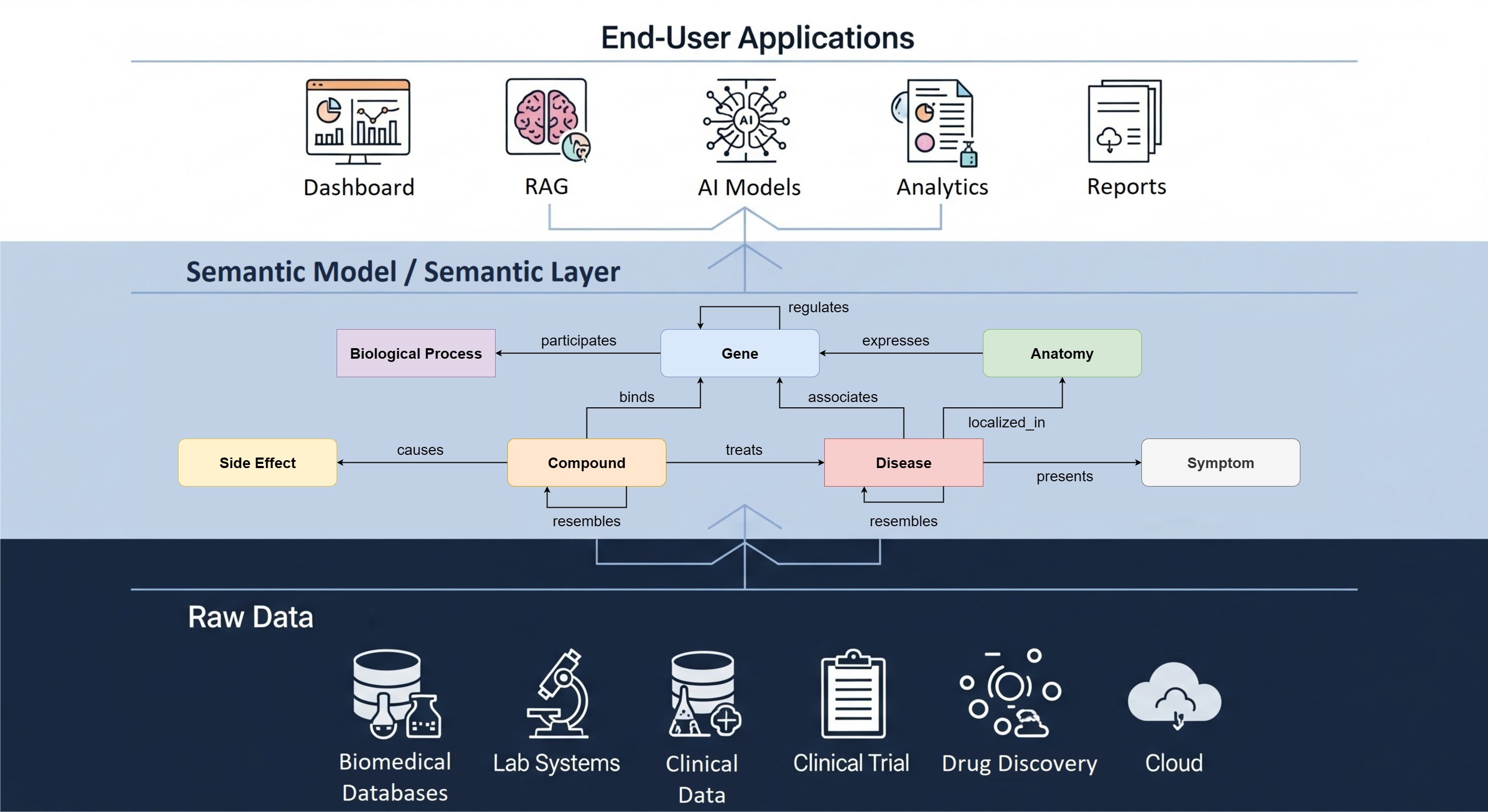
The Hyve has a growing portfolio of open source tools and products that facilitate FAIR research data management throughout academic hospitals. This ranges from using Natural Language Processing(NLP) for medical records processing to building translational datawarehouses with a focus on biomedical research or epidemiology. In the picture below you can see typical data flows throughout a modern university medical center. We use this picture to present a suite of open source tools for data management and analysis.

For Research Data Marts, the main products The Hyve uses are i2b2/tranSMART and cBioPortal (for oncology-focus medical centers or departments). With i2b2/tranSMART, a robust research datawarehouse can be established which exposes a unified patient-centric view of clinical and molecular data for research & analysis purposes. With cBioPortal, this is further enhanced by oncology-specific data visualizations on e.g. mutation profiles of the patient tumour samples.

For Clinical and Administrative Data Marts, this of course overlaps with Research Data Marts but OMOP and its accompanying OHDSI tools are a clinically focused data mart which is specifically built for large scale epidemiology analysis. Besides sources from within the academic hospital, also data from peripheral healthcare organizations (e.g. primary care providers, specialist practices, national registries etc.) can be sourced into an OMOP data mart to analyze the broader care continuum.

For Deep Phenotyping, we have open source product stacks for integrating and curating Electronic Patient Records (Cogstack from KCL, FHIR connectors etc.) that can be connected as inputs for Research / Clinical Data Marts such as tranSMART or OHDSI.
For Biomaterial Sampling, The Hyve can leverage it’s expertise in building custom sample request portals and workflows (e.g. for PALGA and the Hartwig LIMS) based on open source BPM solutions such as Activiti.
For Registry Data, The Hyve is building the Podium open source biobanking research portal in BBMRI. Alternatively, Molgenis or the Montra portal developed in IMI EMIF by University of Aveiro can be used as well, depending on the use case.
For Patient Reported Outcomes and for Sensor Data, The Hyve has the open source RADAR stack that we developed together with King’s College London and other partners in the RADAR-CNS project. This Kafka-based stack offers robust, scalable secure high volume message transport system which can also work over flaky connections such as 4G or Bluetooth. The RADAR Android apps as well as other ResearchKit / ResearchStack apps can be used to directly exchange data with the patients and other care providers.

For Molecular (‘omics’) Data, The Hyve has a range of open source tools available to deploy analysis pipelines and visualization tools via distributed computing, such as Arvados, Molgenis, Cromwell, and iRODS for a more rule based storage and data management system. In order to enhance reproducibility of computations, tools such as GuixSD or NixSD can be used to and to be less dependent on frequently changing orchestration stacks such as Docker.

For Data Querying & Visualization, besides the built in functionalities of tools such as i2b2, tranSMART, cBioPortal and OHDSI, and command line tools and scripting languages such as R and Python, an increasingly popular tool that we use is the Jupyter scientific notebook and the JupyterHub cloud-enabled version. By itself, Juypter already supports many scientific compute kernels such as R, Python, Julia etc. Combined with for example Arvados or Apache Spark as purpose built computation backends, and hooked into a research data mart such as tranSMART via the R or Python connector, this is an extremely powerful setup which can handle almost any data integration use case: it can start with routine visualizations pulling data from the research/clinical data mart and progress into custom built scripts coming e.g. in house with public data.

Finally, on the Data Usage side, an important use cases is FAIR Data Publication. For scientific purposes it is critical that published datasets are FAIR: Findable, Accessible, Interoperable and Reusable, both within as well as outside of the hospital. Tools that can help with this are for example CKAN, which can function as an integrated catalog of available datasets both within as well as to the outside. CKAN is an extremely versatile data repository that is used for open data in general, but also well suited for research data. A similar tool is Dataverse, which was created more in a scientific context Both tools have excellent FAIR rating scores out of the box. Also, the FAIR Data Endpoint protocol can be used e.g. on top of tranSMART to directly expose metadata of available curated studies to a FAIRport.

Although of course there is no hospital in the world where all these tools are used in conjunction with each other, over the years The Hyve has used a fair number of combinations of those open source tools to stand up solutions. We are also actively working with umbrella organisations of for example the Dutch and German academic hospitals, to build reference frameworks for combinations of open source tools and commercially available tools for medical informatics purposes.
Find the PDF version of the poster here.
Please contact us to learn more about open source infrastructure for FAIR Research Data Management in academic hospitals.