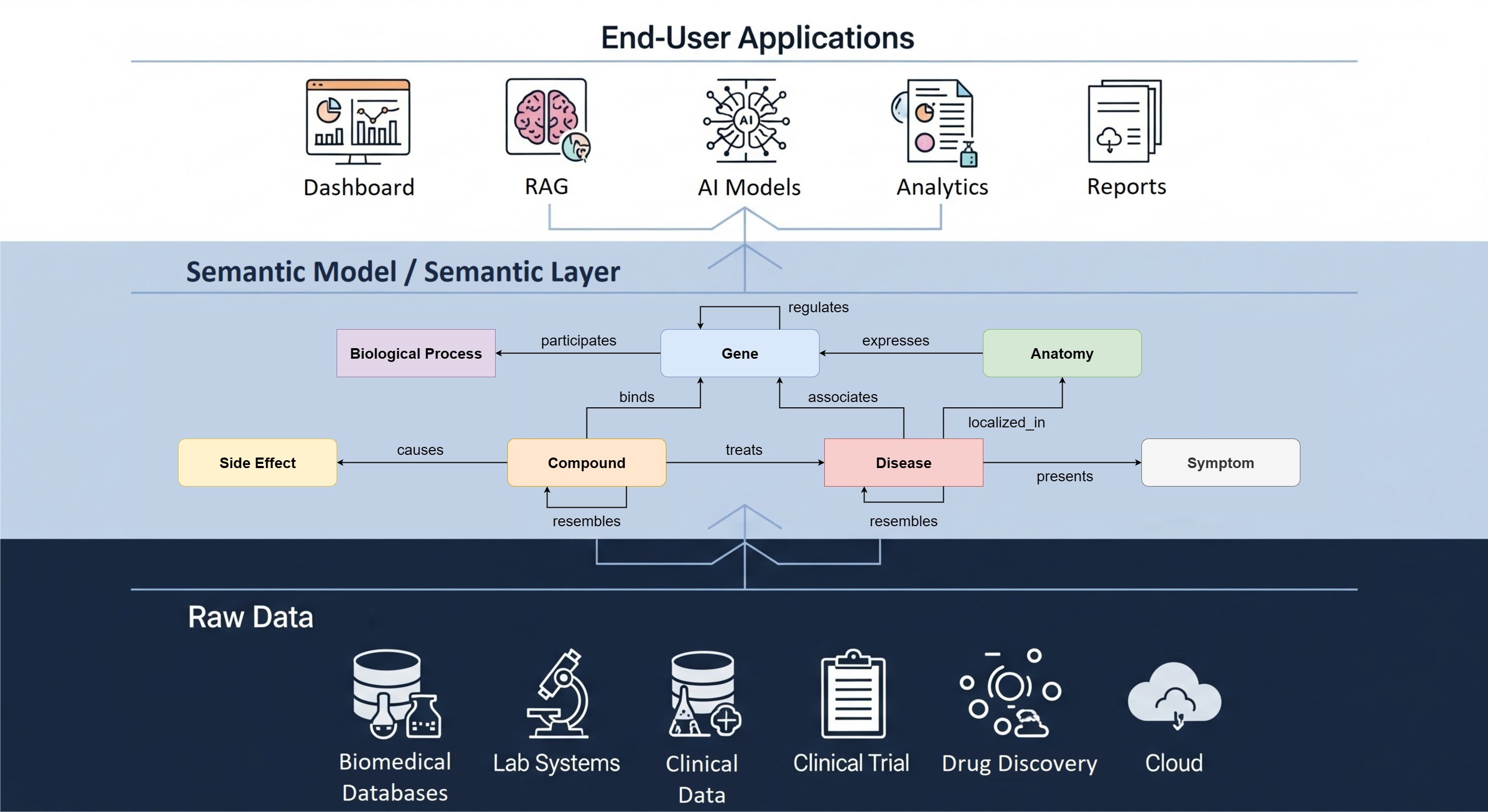
New scientific discoveries increasingly rely on the integration of data from different types of experiments. The Hyve has extensive experience with using the open source tranSMART platform for the storage and analysis of this multi-omics data.
In recent projects we have extended the data types supported by tranSMART, in addition to low dimensional data (demographics, clinical, imaging measurements) and mRNA expression data, with:
- ArrayCGH and miRNA microarray data
- mRNA expression and genomic variants from RNA-Seq and DNA-Seq
- Proteomics and metabolomics data
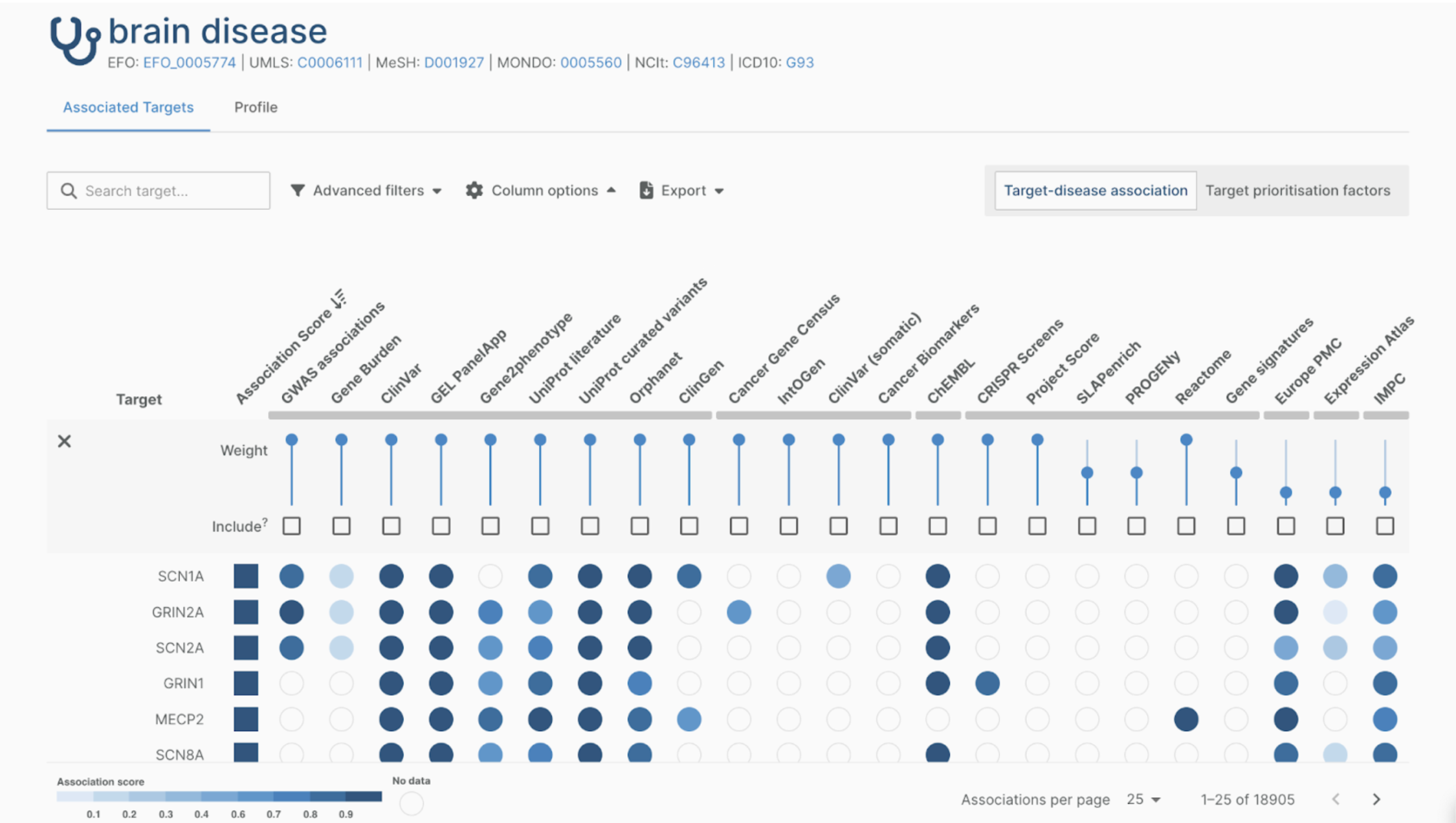
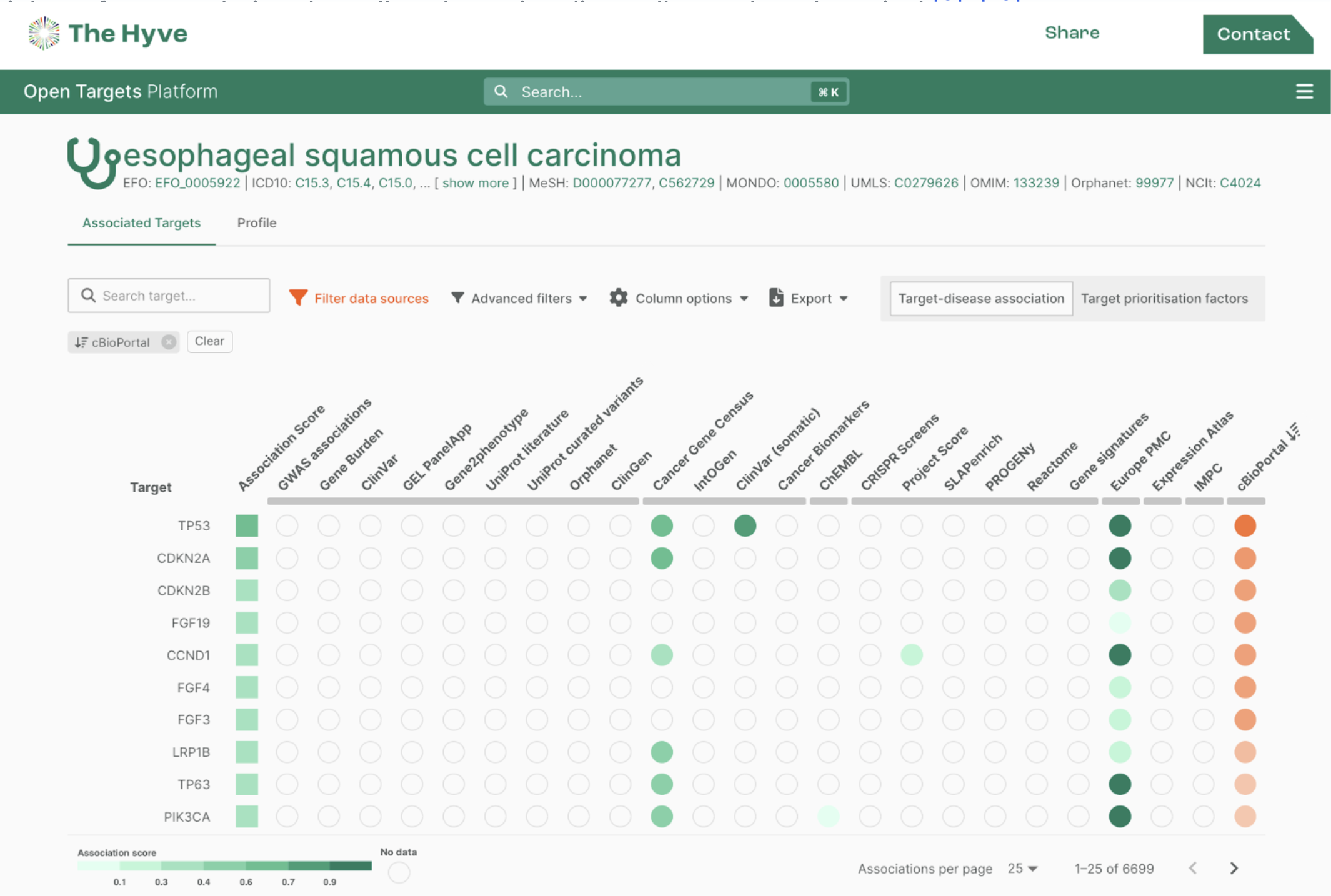
Also we have added multiple analyses to the tranSMART interface which allow the comparison of different data types for the same patient and genes, including the cBioportal OncoPrint. For advanced users and connections to other visualization tools the REST API allows users to retrieve their data from the central tranSMART data warehouse.
Within the Dutch CTMM TraIT project tranSMART functions as the central data warehouse where the data from the multiple data producing domains comes together. To test all these data producing pipelines and the integration the project has brought together data from multiple cell lines from many different modalities. This Cell Line Use Case will soon be made freely available under an open data license.

This poster will be presented at Bio-IT World 2015, one of the largest bioinformatics conferences in the world. Also read our other poster around scalable bioinformatic data storage and analysis with ADAM and Spark.