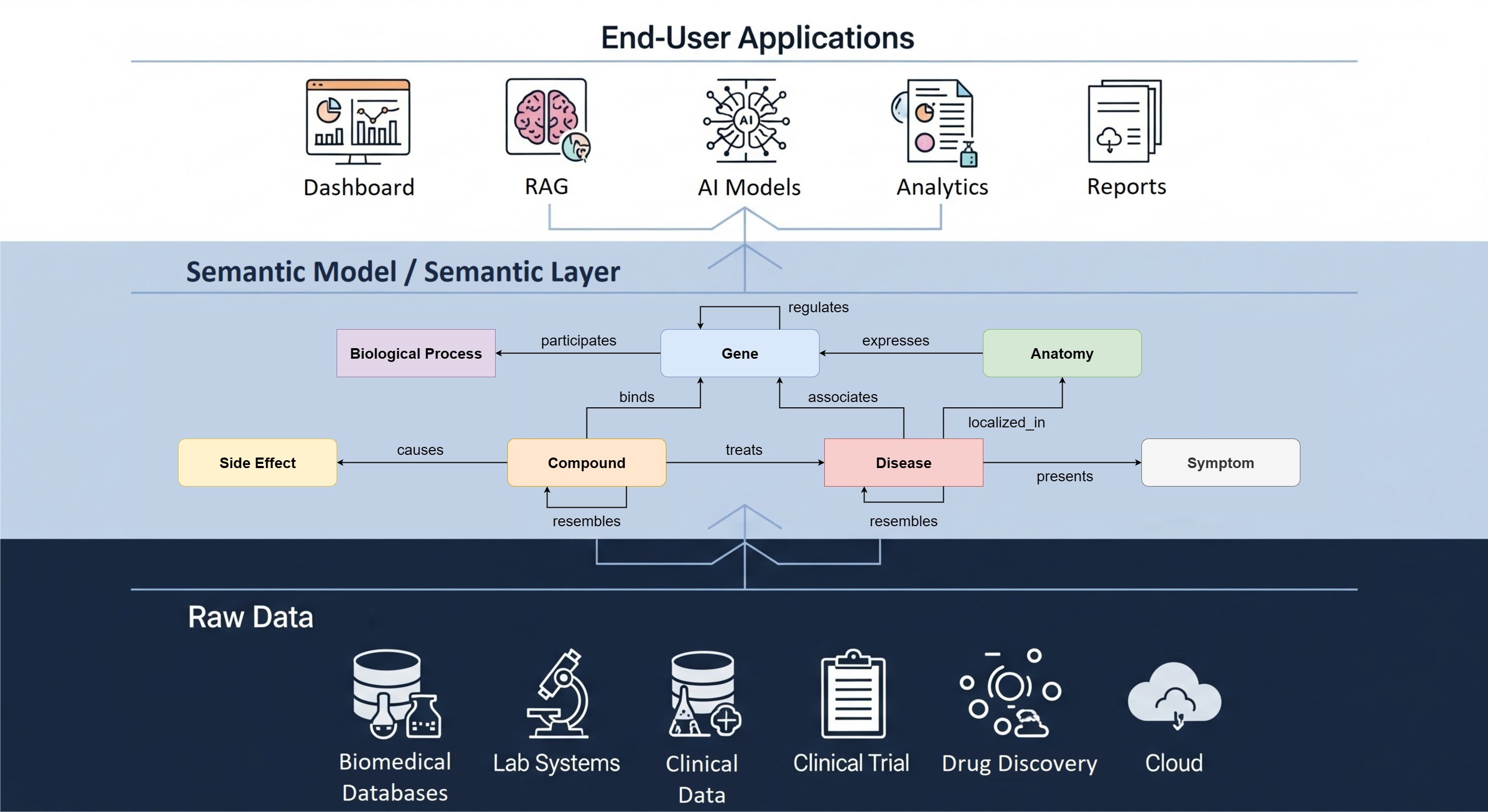
The research data management space has grown and diversified immensely in recent years. The Hyve have been following the developments of this field in the life science industry and academic organizations for a decade now and we were able to collect some interesting insights into open source tools from conferences, customer feedback and our own experience. To get a clear picture of the pros and cons of each tool and being able to advise on the best tool for a particular customer, The Hyve made an investigation last year in which we evaluated a selection of five open source tools that support research data management and identified their strengths and weaknesses. Note that some evaluations might be opinionated and open for discussion.
Evaluation of five open source Research Data Management tools
The team chose to assess five tools: IRODS, Gen3, Fairspace, CEDAR Workbench, and COLID. We selected these tools because we have seen them all used in production environments: we learned about them at conferences, from customer feedback and from our own experience.
The first tool, iRODS (which stands for Integrated Rule-Oriented Data System) is a data virtualization and rule engine framework developed and maintained by the iRODS consortium. iRODS’ data virtualization functionality offers access to distributed storage systems as one familiar file and folder structure, while the rule engine enforces policies and validation, and allows for complicated data management and manipulation workflows. The tool is accessible via command line, though there are graphical user interfaces available too.
The second tool, Gen3, is a platform with a user interface portal for managing, searching, and analyzing large datasets in the cloud. It is being developed by the Center for Translational Data Science at the University of Chicago. The platform powers several public data commons, most notably the NCI’s Cancer Research Data Commons. If you have ever explored data at one of the NIH’s affiliates, the chances are high that you worked with Gen3.
The third tool we evaluated was The Hyve’s own creation: Fairspace. The platform offers a secure portal where researchers can organize their data (files) with rich metadata, all while adhering to the FAIR principles.
The fourth tool, COLID, or Corporate Linked Data, is a data catalog as well as a management system for resolvable identifiers, that is based on FAIR principles/Linked Data principles. It was developed by Bayer and subsequently released as open source. It offers an editor where users can register resources (files, datasets) and a marketplace where these resources can be found based on their attributes.
The fifth tool we evaluated was the CEDAR Workbench developed by the Center for Expanded Data Annotation and Retrieval at Stanford University’s School of Medicine. This tool is used to create metadata submission templates that can subsequently be used by researchers to store and search metadata.
A comparison of the tools’ technical features
We compared and contrasted the five data management tools based on technical criteria (Figure 1), such as code quality and configurability, but also on community criteria, such as activity around the tool, possibilities to get help, and the size and composition of the community.

To be able to compare the strength and weaknesses of each tool, we identified five major application domains (Figure 2): metadata functionality, data management, availability of computational notebook environment, cloud readiness, and analysis/search functionality. We assessed how strong each tool was in each area and plotted the results as a radar-like chart as shown below.

Metadata functionality
Fairspace, COLID, CEDAR, and Gen3 all score high on metadata management. The first three use W3C standards like RDF to define the metadata model, adding to their FAIRness and flexibility, while Gen3 uses its own hierarchical model for metadata management. The metadata model for Fairspace, COLID, and Gen3 need to be defined beforehand, while the CEDAR workbench allows for collaborative evolution of the metadata model and entry forms. The metadata on iRODS objects (files, folders, etc.) consists of free text attribute-value-unit triples that can be searched on in a catalog. Any structure or semantics has to come from custom conventions or implementation of executable rules.
Data management
iRODS is the tool that really shines in data management. It is driven by a rule engine, which lets administrators set up rules to enforce various policies, for example, to enforce fine-grained access policies or to execute complex data manipulation workflows for processing large sequence files or transferring and archiving according to a set schedule or based on events. Both Fairspace and iRODS allow users to organize their data into a familiar file and folder structure. iRODS virtualisation also allows data to be distributed across multiple storage media. Data in Gen3 are managed by referencing files in cloud storage buckets using file nodes in its hierarchical model. Data management in COLID and CEDAR consists purely of providing references to files and datasets. When files or datasets are moved or deleted, the reference should be updated by the user or external application to avoid dangling references. A core part of COLID is its persistent identifier management, which allows users to create a stable, dereferenceable URI, i.e. an URI which stays the same while the underlying reference to the file can be changed, for instance, when the file is moved.
Compute environment
Of the five tools, only two platforms − Gen3 and Fairspace − offer an integrated computational notebook environment. This allows researchers and data scientists to run scripts with transparent access to data and metadata in the system.
Cloud
Many research organizations are moving from on-prem storage and computation to the cloud. Therefore, we wanted to know how cloud-ready each platform was, i.e. how well it utilizes the possibilities that the cloud brings (“cloud-native”). For instance, is it composed of microservices and does it use container orchestration? COLID seems to utilize the cloud setup most, with its microservice architecture, container orchestration, and scaling possibilities. The other tools are suitable for deployment in the cloud and containerization, except Gen 3. Our team did not manage to deploy Gen3 within the one-month project time frame because of its multiple issues.
Analysis
Gen3, COLID, and CEDAR all offer some way of analysis when searching and browsing. With analysis we mean that a user can explore the (meta) data and view summary statistics or graphical representations (charts) of the data. So the user can, for instance, view the fraction of studies in the system that have certain experimental characteristics or cohorts.
Conclusion
When comparing the plots of all five tools (Figure 2), it’s clear that some overlap considerably while others are complementary to each other. COLID and CEDAR are similar: both offer good metadata management, while iRODS offers the best data storage capabilities.
On the domain plot, Gen3 covers most areas well − especially exploring and querying metadata. However, it doesn’t offer really interactive browsing, or changing and modifying the actual data.
The documentation and community aspects of each tool varied as well. iRODS and Gen3 both have very mature documentation with thorough user guides, forums, developer documentations and more. Fairspace and COLID both have significant gaps in their documentation while CEDAR falls in the middle. iRODS, and to a slightly lesser extent Gen3, have a lively and well-developed user community; something that both Fairspace and COLID still need to build.
When it comes to installation, only iRODS and Fairspace are rated as “easy” both locally and in the cloud. Gen3’s installation is rated as “hard” in both areas. COLID had some issues when installing locally.
In summary, the tools are all production ready but each tool has its own best application scenario since all have unique features. Thus it is good to have a clear picture of your specific needs with regard to the five application domains we identified. You can then decide which tool − or maybe two tools − best meets those needs. Of course, The Hyve’s colleagues can support you in this process of identifying your needs and choosing the best tool for your research environment.
We presented this evaluation in November 2022 at the BioIT World Conference & Expo Europe in Berlin and you can access the slides as well.
The Hyve acknowledges support from the European Union’s Horizon 2020 Research and Innovation Programme under grant agreement No 825775, and the Canadian Institute of Health Research under grant agreement No 404896, Common Infrastructure for National Cohorts in Europe, Canada, and Africa (CINECA) project.