Despite notable advancements in fundamental life sciences and biotechnology, the process of discovering and developing drugs continues to encounter obstacles, such as prolonged timelines and substantial costs, averaging 15 years and around US$2 billion for a small-molecule drug. While clinical studies contribute significantly to these costs, the crux of the matter lies in the earlier stages of discovery and preclinical efforts, which account for over 43% of expenses in the pharmaceutical industry. Addressing issues in early discovery, including inadequate target validation or suboptimal drug compounds, is crucial to improving outcomes in preclinical and clinical studies. Streamlining the discovery of diverse and high-quality drug compounds and evaluating their druggability early on could pave the way for more effective, accessible, and safer drugs.
Assessing Druggability and the Druggable Genome Concept
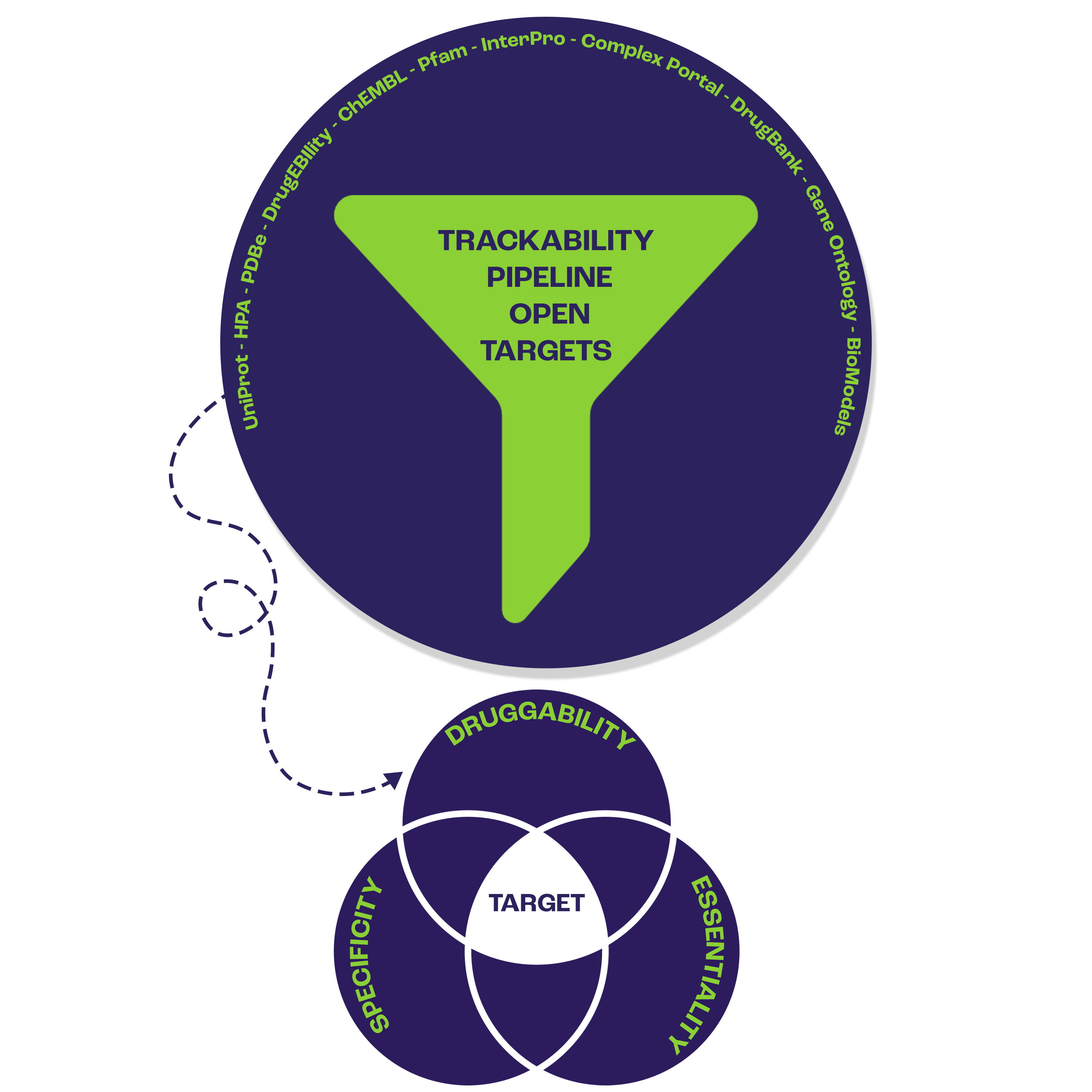
Navigating the complex landscape of drugability involves scrutinizing various molecular characteristics, from structural properties to interactions with cellular components. The challenges in early-stage drug discovery are magnified by the high attrition rate, spanning target selection, hit identification, and lead optimization. For two decades, the concept of the druggable genome has shaped our understanding of target feasibility. Researchers have developed various methods to assess a target's druggability. The Open Targets platform distinguishes itself by integrating extensive datasets, providing researchers with a robust tool to evaluate potential drug targets, including assessment of tractability/druggability, safety and pharmacogenetics. The Tractability Pipeline of Open Targets includes data from UniProt, HPA, PDBe, DrugEBIlity, ChEMBL, Pfam, InterPro, Complex Portal, DrugBank, Gene Ontology, BioModels, as well as data from publications, including a list of druggable genes as per Finan et al.’s Druggable Genome pipeline (2017). This comprehensive set of information equips researchers with valuable insights to make informed decisions in selecting and prioritizing potential drug targets for various therapeutic approaches.
Advancements in Structural Biology and Druggability
Understanding proteins as part of a dynamic ensemble provides a more accurate representation of their behavior, leading to a deeper comprehension of their functions and potential as drug targets. Recent breakthroughs in structural prediction methods, exemplified by AlphaFold2 and RoseTTAFold, have dramatically reduced the time and effort required to obtain structural models. However, proteins are dynamic entities, adopting various structural states crucial for their functions. Experimental techniques such as Nuclear magnetic resonance (NMR), crystallography, and cryo-electron microscopy (cryo-EM) reveal the dynamic nature of proteins, challenging the traditional view of single, static structures.
Open Targets Platform: A Catalyst for Innovation
In the pursuit of overcoming challenges in drug discovery, the Open Targets platform emerges as a beacon of innovation. This collaborative initiative, spearheaded by leading research institutions and pharmaceutical companies, harnesses the power of advanced algorithms and data analytics. By integrating genetic, genomic, and drug-related information, the platform accelerates the identification and validation of promising drug targets. Moreover, the platform has tremendous potential for integrating its data from gene-level to protein residue in a single knowledge graph, facilitated by scalable, automated workflows. Utilizing graph-based AI methods as discussed previously, particularly with advancements like structural prediction methods, could expertly navigate such a knowledge graph, aiding in the selection of future targets. At The Hyve we are specialized in tailoring Open Targets for customers, offering a customized platform instance that aligns with their specific requirements.


